Definition
Gene Ontology (GO) is the world’s largest source of information on the functions of genes. This knowledge is both human-readable and machine-readable, and is a foundation for computational analysis of large-scale molecular biology and genetics experiments in biomedical research.
The Core Problem
Scientists worldwide study genes using different terminology for identical biological concepts. One researcher might refer to “TNF-alpha,” another uses “tumor necrosis factor,” and a third simply writes “TNF.” This inconsistency creates chaos when attempting to integrate research findings or compare data across studies.
The mission of the Gene Ontology Consortium is to provide a comprehensive and up-to-date computational model of the current scientific understanding of the functions of gene products. GO encompasses all levels of biological systems, from molecular activities to complex cellular and organismal-level networks, providing uniform descriptors applicable to gene products across the entire tree of life.
Historical Origins
Founded in 1998, the project began as a collaboration between three model organism databases: FlyBase (Drosophila), the Saccharomyces Genome Database (SGD) and the Mouse Genome Database (MGD). The consortium has since expanded to incorporate databases representing the world’s major repositories for plant, animal, and microbial genomes.
Two Core Components
The Ontology: Structured Knowledge
The network of biological classes describing the current best representation of the “universe” of biology: the molecular functions, cellular locations, and processes gene products may carry out.
Each term in GO contains:
- A unique identifier (e.g., GO:0006954)
- A precise definition
- Relationships to other terms
- Evidence codes linking to scientific literature
The Annotations: Gene-Term Associations
Statements, based on specific, traceable scientific evidence, asserting that a specific gene product is a real exemplar of a particular GO class.
As of October 2024, the GO includes experimental findings from over 180,000 published papers, representing over 1,000,000 experimentally-supported annotations.
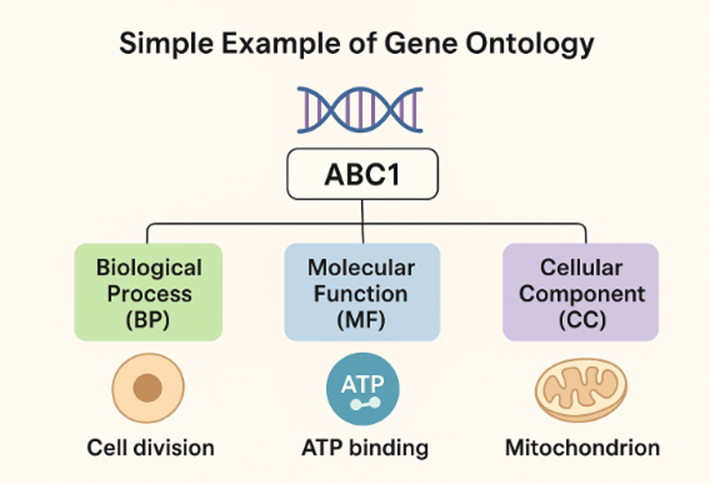
Three Ontological Domains
The GO is organized in three aspects: Molecular Function (MF), Cellular Component (CC), and Biological Process (BP).
Molecular Function (MF): Activities performed at the molecular level, such as “catalysis” or “transcription regulator activity”
Cellular Component (CC): Locations where gene products perform their functions within the cell
Biological Process (BP): Larger biological programs accomplished by multiple molecular activities
The three GO aspects are disjoint, meaning that no is_a relation exists between terms from different ontology aspects. However, other relationships such as part_of and occurs_in can operate between terms from different GO aspects.
Structural Architecture: Directed Acyclic Graph
The structure of GO is a Directed Acyclic Graph (DAG), where the terms are the nodes and the relations among terms are the edges.
Gene Ontology is organized not as a simple hierarchy, but as a directed acyclic graph. This structure reflects the reality that biological categories often overlap. A GO term can belong to multiple higher-level categories, inheriting traits from each.
Key Structural Properties
The edges are directed—there is a source and a destination for each edge. In gene ontology, the source is referred to as the parent term and the destination as the child term. Unlike a general graph, a DAG does not have cycles, meaning one cannot complete a loop by following directed edges.
GO is hierarchical, with child terms being more specialized than their parent terms, but unlike a strict hierarchy, a term may have more than one parent term.
Common relationships include:
- is_a: Child term is a more specific instance of parent term
- part_of: Component relationship between terms
- regulates: Regulatory relationships between processes
Scale and Impact
As of July 2019, the GO contains 44,945 terms with 6,408,283 annotations to 4,467 different biological organisms. There is a significant body of literature on the development and use of GO, and it has become a standard tool in the bioinformatics arsenal.
The GO knowledgebase plays an essential role in supporting biomedical research and has been cited in tens of thousands of scientific studies.
Primary Applications
The most common use of GO annotations is for interpretation of large-scale molecular biology experiments, to gain insight into the structure, function, and dynamics of an organism. Gene Ontology enrichment analysis is used to discover statistically significant similarities or differences under alternate controlled experimental conditions.
Enrichment Analysis
One of the main applications of GO is the identification of over- or under-represented GO-terms for a specified gene list (as a result from identifying differentially expressed genes) utilizing a hypergeometric test (also known as Fisher’s exact test).
This approach helps categorize genes into functional groups, highlighting enriched biological processes, molecular functions, or cellular components in experimental datasets.
Cross-Species Comparison
GO is increasingly used to compare gene products across species and to find functional patterns within groups of genes. The gene ontology hierarchy encapsulates functional homology between genes, which may not be evident from other methods of interspecies gene comparison, such as sequence alignment.
Annotation Evidence
Annotations from automated processes are given the code Inferred from Electronic Annotation (IEA). As of July 2, 2019, only about 30% of all GO annotations were inferred computationally. As these annotations are not checked by a human, the GO Consortium considers them to be marginally less reliable.
Each annotation includes:
- Gene product identifier
- GO term
- Evidence code documenting experimental or computational support
- Reference to published research
- Date and curator information
Tools and Access
The GO Consortium develops and supports two tools: AmiGO and OBO-Edit. AmiGO is a web-based application that allows users to query, browse, and visualize ontologies and gene product annotation data. OBO-Edit is an open source, platform-independent ontology editor developed and maintained by the Gene Ontology Consortium.
Available Resources
Official Data Sources:
- Website: https://geneontology.org
- The Gene Ontology Consortium provides data in multiple formats, the most common being OBO and OWL for the ontology structure, and GAF for annotations.
Analysis Tools: Third-party tools including DAVID, Enrichr, PANTHER, clusterProfiler, and numerous others support GO-based functional analysis.
Dynamic Evolution
The GOC has expanded areas of the ontology such as cilia-related terms, cell-cycle terms and multicellular organism processes. The consortium has implemented new tools for generating ontology terms based on logical rules using templates.
Members of the GO Consortium continually work collectively, involving outside experts as needed, to expand and update the GO vocabularies. This ensures the ontology remains current as biological understanding advances.
Funding and Governance
The GO Consortium is funded by the National Human Genome Research Institute (US National Institutes of Health), grant number HG012212, with co-funding by NIGMS.
Practical Example
Query: “What genes are involved in inflammatory response?”
Without GO: Manually search millions of papers, handle inconsistent terminology, miss synonyms, compile fragmented lists.
With GO:
- Query GO:0006954 (inflammatory response)
- Retrieve all genes annotated to this validated term
- Access curated information with evidence codes
- Explore hierarchical relationships to parent/child processes
- Discover related pathways and molecular functions
Real-World Research Applications
Gene Ontology supports multiple research workflows:
Biomarker Discovery: Identifying genes linked to specific diseases or conditions
Drug Target Identification: Finding genes involved in disease pathways for therapeutic intervention
Pathway Analysis: Understanding biological mechanisms underlying experimental observations
Machine Learning: Providing structured biological knowledge for AI model training and validation
Technical Implementation
GO has a hierarchical structure in the form of a directed acyclic graph (DAG), which means that GO-terms are dependent on each other. This interdependence requires specialized tools and statistical methods for proper analysis.
The GO database consists of a MySQL database that captures GO content and a Perl object model and Application Programmer Interface (API) to simplify database access and help programmers write tools that use GO data.
Community Contribution
The GO effort is greatly enriched by input from its user community. Comments and suggestions for changes and updates to the ontologies can be submitted via a GO project page at the SourceForge site.
The collaborative nature of GO ensures continuous improvement driven by the broader research community’s needs and discoveries.
Significance for Biomedical AI
For computational biology and AI applications in life sciences, Gene Ontology provides:
Standardized terminology: Eliminates ambiguity in biological knowledge representation
Structured relationships: Enables graph-based reasoning and multi-hop inference
Validated knowledge: Expert-curated annotations reduce hallucination risk in AI systems
Machine-readable format: Supports integration with RAG systems, knowledge graphs, and semantic search
Evidence tracking: Every annotation traces to peer-reviewed literature, enabling explainable AI
Gene Ontology transforms scattered biological knowledge into a computationally accessible framework, making it indispensable for modern biomedical research and AI-powered discovery systems.

Leave a Reply